Filter datasets
About this tool
This tool provides an opportunity to quickly analyze mass spectrometric data and to store the results. The data are submitted as CSV or XLSX files, either containing intensities of individual proteins or logP/LFC values calculated elsewhere. The tool generates LFC/LogP volcano plot with a corresponding data table, performs multiple pathway analyses and offers an opportunity co compare datasets with each other.
Instructions
Creating a user and signing in
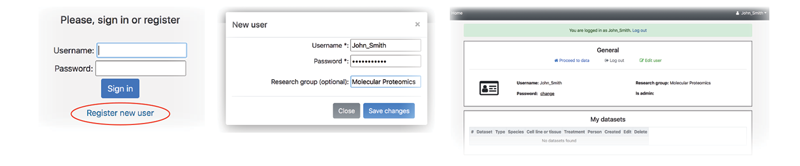
In order to get access to personal datasets and full analysis possibilities, it is advisable to log in with personal credentials. If you have not registered yet, please, create a new user by clicking the corresponding button. After successful sign in, you will be forwarded to your personal page where you can manage your datasets and other information.
Uploading CSV files and creating new datasets
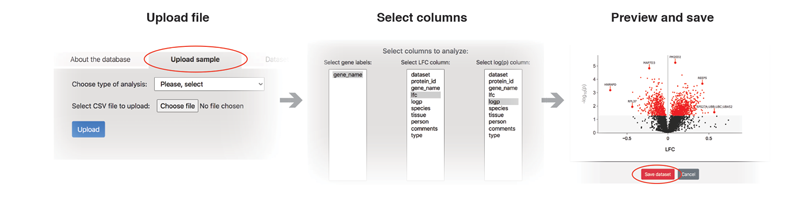
The "Upload file" link is used to upload and analyze a new file. It is very important to choose a right analysis type, as this will define the technical details on how the data will be analyzed. After the file is submitted, a new page will appear where one can choose relevant columns from the file. After the submission, a preview of the volcano plot will be created. If the result looks acceptable, the dataset needs to be saved for subsequent analysis.
Dataset analysis
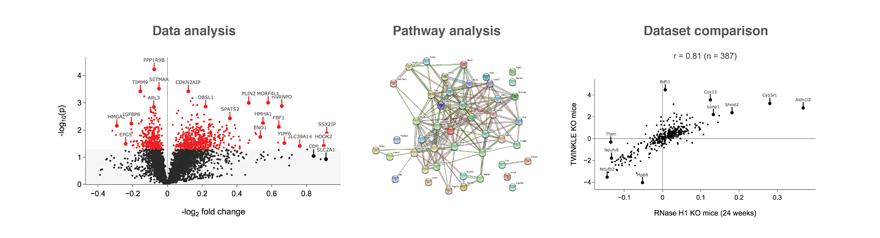
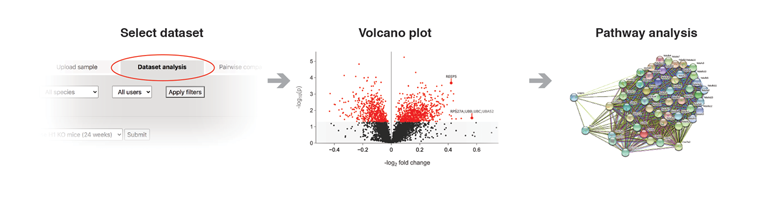
To analyze a saved dataset, please, go to the "Dataset analysis" button. Only the datasets accessible by the current user will be listed. Select a dataset and press "Submit". After a few seconds, the data will be displayed in several blocks. Besides the volcano plot and the corresponding data table, STRING and Reactome analyses will be performed.
Comparing multiple datasets
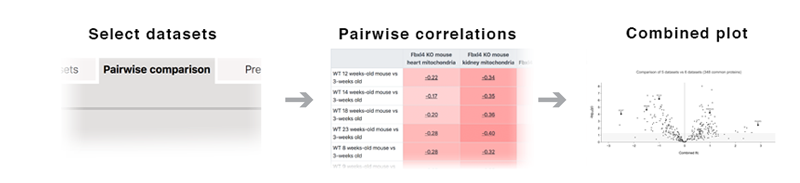
Several (two or more) datasets can be compared under the "Multiple comparison" tab. Selected datasets will be compared to each other and the results displayed as heat maps and/or scatterplots comparing individual proteins and changes in pathways.
Grouped dataset comparison
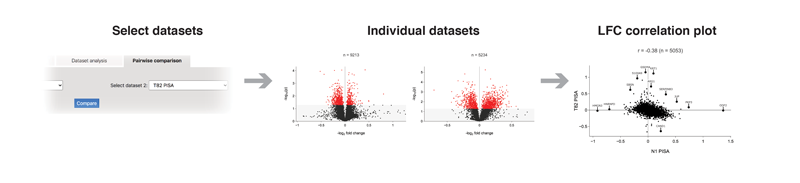
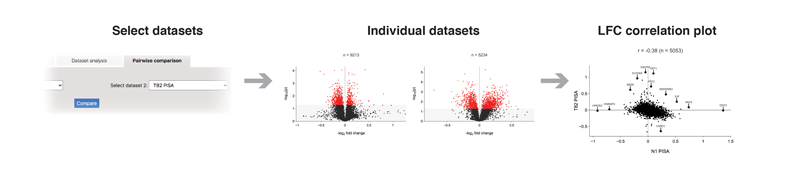
The "Pairwise comparison" tab allows comparing two groups of datasets against each other. LFC values for each of the proteins will be combined and represented as a comparison scatter plot and as a combined volcano plot.
Prey analysis
Here you can investigate the behavior of a selected protein across datasets, e.g. to find out under which conditions it is up- or downregulated. Moreover, this type of analysis offers possibility to compare behavior of two proteins and to find other proteins that follow the same expression pattern.
Contact
In case of any technical issues or other queries, please, contact oleksandr.lytovchenko@ki.se.
1. Choose the right analysis type: if you have pre-calculated (log) fold-change and LogP values, use the
"from (L)FC and (log)P" option, otherwise use "calculate from intensities".
2. Upload data file in any suitable format (xlsx, csv, txt, odt). If the file is not read properly, you
can try to save it in a different format and re-submit.
For example, you can read
here
how to convert Excel files to csv.
To analyze one of the saved datasets, select if from the drop-down list and press "Display".
Private and public datasets are listed separately in an alphabetic order.
To reduce number of displayed datasets, filters can be applied by selecting corresponding options and
pressing "Apply filters".
To display all datasets again, press "Reset".
Two or more datasets can be compared to each other. If two datasets are selected, LFC comparison plot
will be displayed in addition to other analyses.
Datasets of interest can be selected in the multiple selection list and moved to the "Order of selected
datasets" list in the desired order. Datasets can be removed from the list using the left arrow button.
Use this tab to analyze behavior of individual proteins across datasets.
The results will be displayed as LFC/LogP values for the selected gene in each dataset where it has been
detected.
Two proteins can be also compared to each other to see if they behave similarly across datasets.
Use this tab to perform pairwise analysis of two groups of datasets (or two individual datasets).